15 Integration Using Monte Carlo and Markov Chain Techniques
In the previous chapters, we used representative samples of posterior distributions to answer questions using indicator functions. While we relied on these methods to make probabilistic statements, you may have wondered how and why they work. In this chapter, we delve deeper into the computational mechanisms behind Bayesian inference and explore why representative samples are critical for obtaining accurate posterior distributions in practical problems. By examining the inner-workings of integration methods and their approximations, we will actually find a deeper understanding of the underpinnings of Bayesian inference and how it enables us to make robust probabilistic statements using numpyro.
15.1 Probabilistic Statements Are Just Integrals
For the moment, let’s assume we have a concise way of representing a joint posterior distribution - let’s call this distribution
When we tell narrative stories about the plausibility of potential model configurations, those stories are encoded in density
Integrals, like the above, cannot be evaluated analytically for most of the practical problems data analysts face, hence the data storyteller will resort to numerical methods for approximating answers to the above. The approximations will come in the form of output from a class of algorithms known as Monte Carlo methods. We introduce these methods using an illustrative example.
15.1.1 An Example Target Distribution
For our example, let’s imagine a game where you will win money based on the output of two random events. The first random event is that you will get a chance to spin the below spinner.
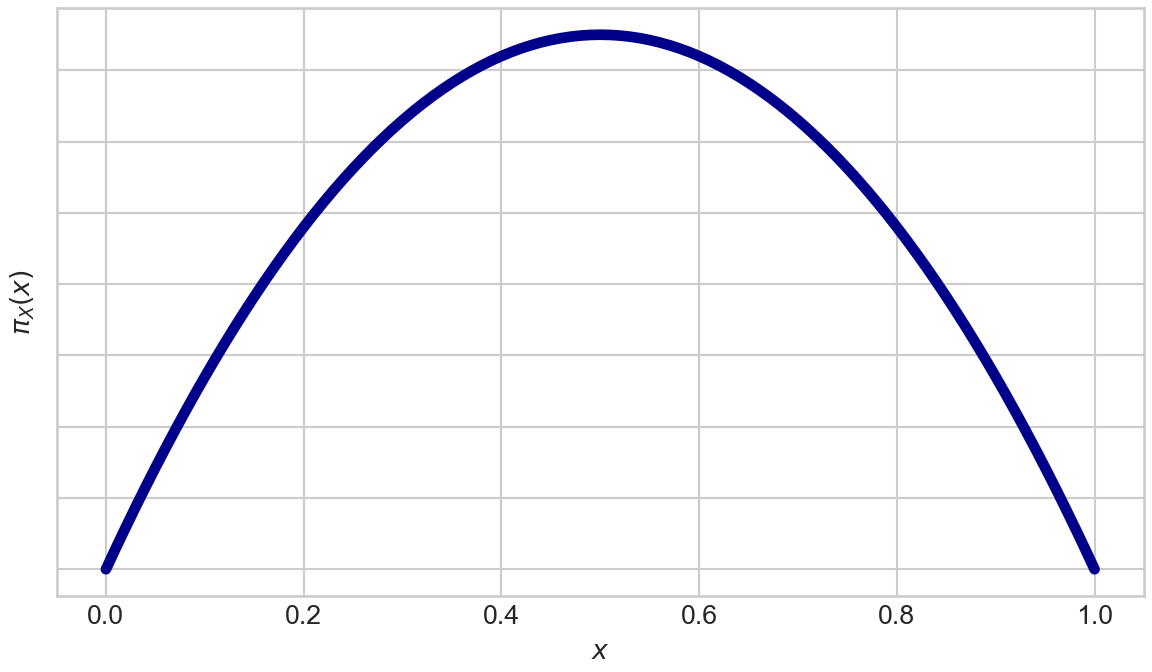
The number the spinner ends up pointing to, a decimal between 0 and 1, is your win multiplier. Let’s call it
We graph the above density function with the following code:
import matplotlib.pyplot as plt
plt.style.use("seaborn-whitegrid")
def pi_X(x):
if x >= 0 and x <= 1:
return 6*x*(1-x)
else:
return 0
x = [i/100 for i in range(101)]
y = [pi_X(i) for i in x]
fig, ax = plt.subplots(figsize=(6, 3.5), layout='constrained')
ax.plot(x, y, color='darkblue', linewidth=4)
ax.set(xlabel=r'$x$', ylabel=r'$π_X(x)$')
ax.set_yticklabels([])
plt.show()From Figure 15.2, we see outcomes near 0.5 that are associated with the “higher friction zone” shown in Figure 15.1 are truly more probable than outcomes near 0 or 1.
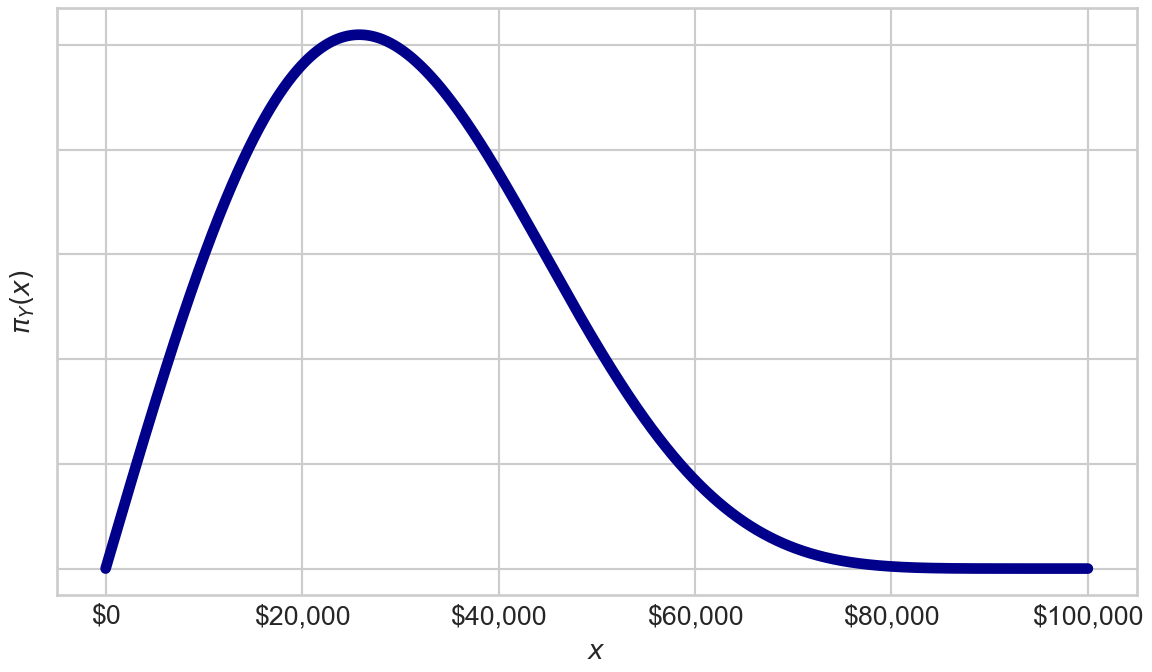
Additionally, let’s assume the probability density function for the day’s maximum win amount is also known:
This is actually derived from a Kumaraswamy distribution, but let’s ignore the obscure distribution name and just plot the density of
from matplotlib.ticker import FuncFormatter
def pi_Y(y):
kumaRealization = y / 100000
jacobian = 1 / 10**5
if kumaRealization >= 0 and kumaRealization <= 1:
return 2*8*kumaRealization*(1-kumaRealization**2)**7*jacobian
else:
return 0
def dollar_format(x, pos):
"""Format y-axis tick labels in dollar format"""
return '${:,.0f}'.format(x)
x = [i for i in range(100001)]
y = [pi_Y(i) for i in x]
fig, ax = plt.subplots(figsize=(6, 3.5), layout='constrained')
ax.plot(x, y, color='darkblue', linewidth=4)
ax.set(xlabel=r'$x$', ylabel=r'$π_Y(x)$')
ax.xaxis.set_major_formatter(FuncFormatter(dollar_format))
ax.set_yticklabels([])
plt.show()Figure 15.3 shows players can win up to $100,000, but more plausible values of max winngins are typically less than $50,000. Thus, while the game designers might advertise you can win up to $100,000 (i.e.
Let’s answer this by expressing the game in the language of Equation 15.1. Let
The function,
I do not know about you, but I see that integral and I do not want to attempt it analytically. While it’s not impossible to compute the given integral analytically, it would require significant effort and advanced mathematical knowledge. Numerical methods provide a more practical approach for obtaining an estimate of the integral. The numerical way is the easier way.
Practical Bayesian problems often involve complex models with intractable posterior distributions that cannot be computed analytically. Therefore, numerical approximation methods such as Markov chain Monte Carlo (MCMC) are commonly used to sample from the posterior and approximate the required integrals. MCMC can be computationally intensive, but it is often the most practical approach for approximating the posterior distribution in complex Bayesian models.
15.1.2 Monte Carlo Integration
One way to numrically tackle analytically-challenging integrals is to approximate them using Monte Carlo integration. Recall from calculus (or watch the below video) that an integral, like Equation 15.2, can be approximated as a summation of small regions whose total becomes a volume measurement over the region of integration.
So, thinking of Equation 15.2 as a volume of some shape, it is approximated by summing smaller shapes that consist of length
Defining
(see this article for the more general formula). Intuitively, think of each summand as the value of the integrand at a specific
In Python, we pursue
import pandas as pd
N = 1000 ## sample using 1000 points
gridDF = pd.DataFrame({'winningsMultiplier': np.random.rand(N),
'maxWinnings': 10**5 * np.random.rand(N)})
fig, ax = plt.subplots()
ax.scatter(gridDF['winningsMultiplier'], gridDF['maxWinnings'], color='darkblue')
ax.yaxis.set_major_formatter(FuncFormatter(dollar_format))
# Set the axis labels and title
ax.set_xlabel('Winnings Multiplier')
ax.set_ylabel('Max Winnings')
ax.set_title('Randomly-Spaced Sampling Points')
plt.show()And we can then add information about evaluating the integrand of Equation 15.2 or equivalently the summand of Equation 15.3 at each of the 1,000 grid points.
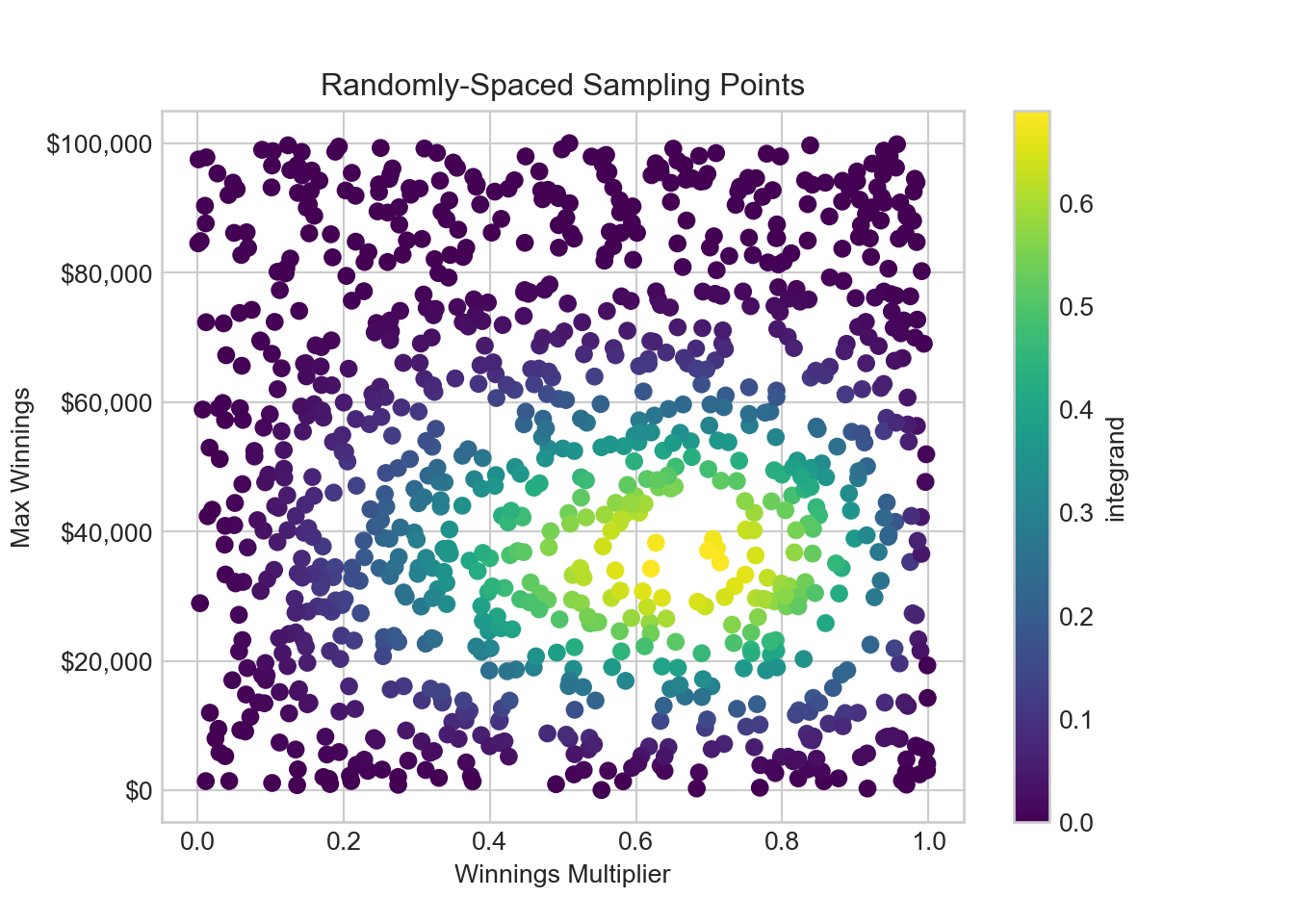
# Define the integrand function
def integrandFun(x, y):
return 3 * x**2 * y**2 / (3125 * 10**5) * (1 - x) * (1 - y**2 / 10**10)**7
# Add a column to the DataFrame using the above function
gridDF['integrand'] = gridDF.apply(lambda row: integrandFun(row['winningsMultiplier'], row['maxWinnings']), axis=1)
fig, ax = plt.subplots()
winPlot = ax.scatter(gridDF['winningsMultiplier'], gridDF['maxWinnings'], c=gridDF['integrand'], cmap='viridis')
ax.yaxis.set_major_formatter(FuncFormatter(dollar_format))
# Set the axis labels and title
ax.set_xlabel('Winnings Multiplier')
ax.set_ylabel('Max Winnings')
ax.set_title('Randomly-Spaced Sampling Points')
fig.colorbar(winPlot, label='integrand')plt.show()From Figure 15.7, we see that the largest contributors to
In code, our estimate of expected winnings turns out to be:
expectedWinnings = 10**5 * np.mean(gridDF['integrand'])
print("Expected Winnings: " + dollar_format(expectedWinnings, None))Expected Winnings: $15,194Your calculation for expected winnings may be different than mine (i.e. due to inherent randomness in Monte Carlo integration). However, asymptotically if we were to increase
gridDF.sample(n=10) winningsMultiplier maxWinnings integrand
702 0.834962 3261.267441 0.011661
204 0.019871 43354.324872 0.001626
985 0.157636 39152.398135 0.096104
739 0.794737 81238.281805 0.004317
892 0.601258 86124.118429 0.000787
261 0.969923 4789.926428 0.006133
500 0.305750 28410.414633 0.279014
850 0.038085 33348.562892 0.006526
667 0.114836 65197.370747 0.009890
143 0.750703 9848.917400 0.12219915.1.3 More Efficient Monte Carlo Integration
If the highest summands/integrands sway our expected estimates much more dramatically than all of the close-to-zero summands, then our estimation error could be reduced more efficiently if our estimation method spends more time sampling from high summand values and less time sampling at near-zero summands. One way to accomplish this is to use a non-uniform sampling of points known as importance sampling; sample the far-from-zero summands more frequently and the close-to-zero summands less frequently. In doing this, each point can no longer be weighted equally, but rather our estimate gets adjusted by the probability density function of drawing the non-uniform sample (see Wojciech Jarosz (2008)). Labelling the sampling probability density function
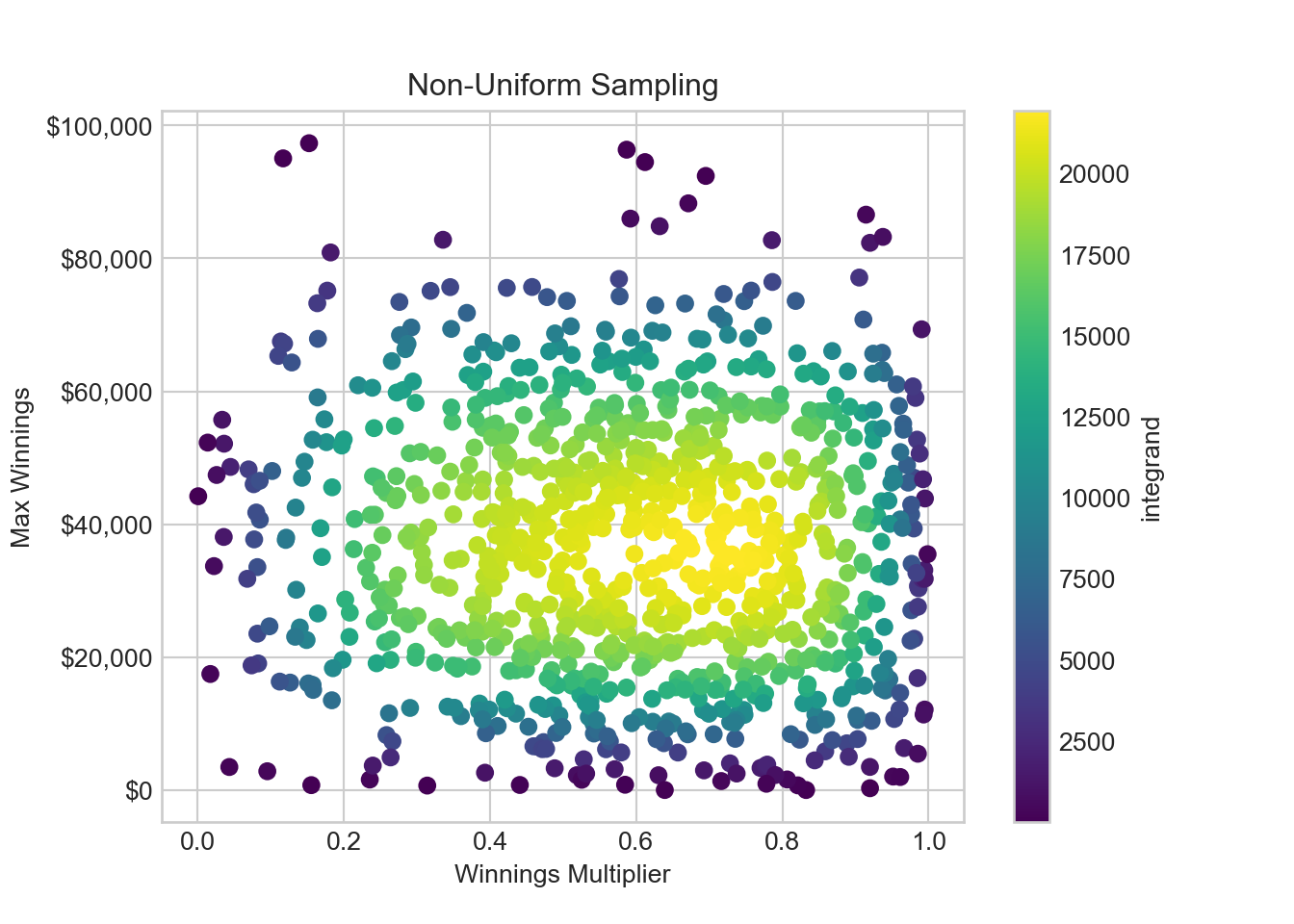
from scipy.stats import truncnorm
## bounds of distributions are in sd's, not actual bounds
xSamplingDist = truncnorm((0 - 0.65) / 0.3, (1 - 0.65) / 0.3, loc=0.65, scale=0.3) # winnings winningsMultiplier
ySamplingDist = truncnorm((0 - 35000) / 20000, (100000 - 35000) / 20000, loc=35000.0, scale=20000.0) # maxWinnings
# sample from the distributions
np.random.seed(123) # for reproducibility
n = 1000 # sample size
winningsMultiplier = xSamplingDist.rvs(n)
maxWinnings = ySamplingDist.rvs(n)
integrand = integrandFun(winningsMultiplier, maxWinnings) / (xSamplingDist.pdf(winningsMultiplier) * ySamplingDist.pdf(maxWinnings))
# create a scatter plot
fig, ax = plt.subplots()
winPlot = ax.scatter(winningsMultiplier, maxWinnings, c=integrand, cmap='viridis')
# set the axis labels and title
ax.yaxis.set_major_formatter(FuncFormatter(dollar_format))
ax.set_xlabel('Winnings Multiplier')
ax.set_ylabel('Max Winnings')
ax.set_title('Non-Uniform Sampling')
fig.colorbar(winPlot, label='integrand')plt.show()With this configuration, Figure 15.8 shows the sweetspot of x-y coordinates near 0.65 and $35,000 is now sampled much more intensely. Additionally, we see that our re-weighted integrand leads to each and every sampled point having more similar integrand values. This is a nice feature - each sample makes a more equal and significant contribution to the final estimate.
15.1.4 Estimate Sensitivity to Choice of Sampling Distribution
In the above examples of Monte Carlo integration, we approximated an integral that I was too intimidated by to attempt analytically. They gave us useful and consistent information, namely that expected winnings are in the neighborhood of $15,000 give or take a few thousand dollars. While the technique promises convergence to an exact solution, it is unclear how many samples are needed to get a level of approximation that we are comfortable with. I like to simply run the algorithms a few times and see how consistent the answers are from one run to the next. This yields an idea of how accurate the estimates might be.
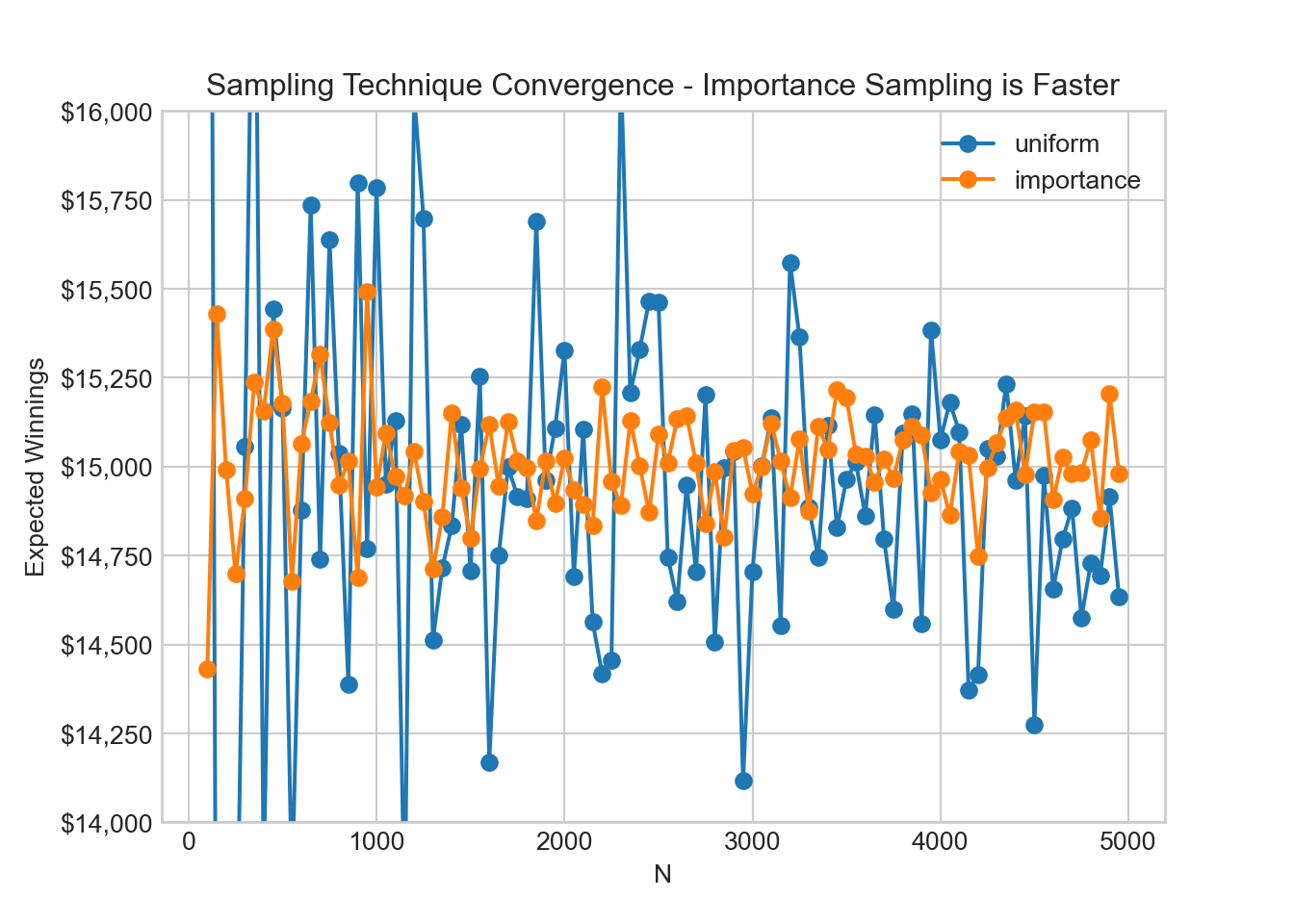
# comparison of convergence
N_values = np.arange(100, 5000, step=50)
sim_results = []
def integrandFun2(x, y):
return integrandFun(x, y) / (xSamplingDist.pdf(x) * ySamplingDist.pdf(y))
for N in N_values:
# technique 1 grid - uniform sampling
winningsMultiplier = np.random.rand(N)
maxWinnings = 10**5 * np.random.rand(N)
integrand_unif = integrandFun(winningsMultiplier, maxWinnings)
expWinnings_unif = 10**5 * np.mean(integrand_unif)
sim_results.append((N, 'uniform', expWinnings_unif))
# technique 2 grid - importance sampling
x_samples = truncnorm.rvs((0 - 0.65) / 0.3, (1 - 0.65) / 0.3, size=N, loc=0.65, scale=0.3)
y_samples = truncnorm.rvs((0 - 35000) / 20000, (100000 - 35000) / 20000, size=N, loc=35000, scale=20000)
integrand_imp = integrandFun2(x_samples, y_samples)
expWinnings_imp = np.mean(integrand_imp)
sim_results.append((N, 'importance', expWinnings_imp))
# plot the results
fig, ax = plt.subplots()
for label in ['uniform', 'importance']:
x, y = zip(*[(N, expWinnings) for N, method, expWinnings in sim_results if method == label])
ax.plot(x, y, label=label, marker='o')
ax.set_xlabel('N')
ax.set_ylabel('Expected Winnings')
ax.set_title('Sampling Technique Convergence - Importance Sampling is Faster')
ax.set_ylim(14000, 16000)ax.yaxis.set_major_formatter(dollar_format)
ax.legend()
plt.show()As seen from the above figure, as we increase the number of sample points,
15.2 Sampling Efficiency
As one gets to higher dimensional problems, beyond two variables like
Recall the integral we want to approximate has a value accumulated over a volume of probability space
Where the expectation
When using importance sampling to approximate this integral, we made a grid of points using probability density
where
Previously, we used Monte Carlo integration which is equivalent to plugging a uniform
The mathematical ideal is to minimize the variance, denoted
where the first simplification step is due to drawing independent samples of
The math reveals insight to reduce our estimator variance. Either we increase
15.3 Markov Chain Monte Carlo
Since our goal is speed, we need to aim for sampling from
Michael Betancourt (2018) justifies this simplification well:
“In practice we are often interested in computing expectations with respect to many target functions, for example in Bayesian inference we typically summarize our uncertainty with both means and variance, or multiple quantiles. Any method that depends on the specifics details of any one function will then have to be repeatedly adjusted for each new function we encounter, expanding a single computational problem into many. Consequently, [we assume] that any relevant function of interest is sufficiently uniform in parameter space that its variation does not strongly effect the integrand.”
15.3.1 The Mathematical Tricks Used in MCMC
The key idea, widely used in practice, is to get
where to estimate
The most-notable class of algorithms that support getting samples from
Surprisingly, the algorithms that approximate drawing independent samples from
The algorithms yield samples as if they were drawn from
A properly constructed transition function
- Detailed Balance: The probability of being at any one point
- Ergodicity: Each sample
Hastings (1970) then tells us to separate the transition density
- Pick any starting point
- Propose a point
- Calculate the acceptance probability of the proposed jump using the Metropolis acceptance criteria,
- Set
- Repeat steps 2-4 until convergence (to be discussed later).
15.3.2 Demonstrating the Metropolis Sampler
Recall our spinner example where want to sample a joint distribution of a winnings multiplier
where we are assuming we do not have a good way to sample from these two densities. Let
15.3.3 Step 1: The Starting Point
# initial point - pick extreme point for illustration
qDF = pd.DataFrame({
'winningsMultiplier': [0.99],
'maxWinnings': [99000.0]
})15.3.4 Step 2: Propose a point to jump to
from numpy.random import default_rng
rng = default_rng(54321)
rng = default_rng(123)
jumpDist = rng.multivariate_normal(np.zeros(2), np.diag([0.1**2, 200**2]))
q_current = qDF.iloc[0].to_numpy()
q_proposed = q_current + jumpDist15.3.5 Step 3: Calculate acceptance probability
# density of q when params passed in as array
def pi_Q(q):
return pi_X(q[0]) * pi_Y(q[1])
acceptanceProb = min(1, pi_Q(q_proposed) / pi_Q(q_current))
print(acceptanceProb)115.3.6 Step 4: Add point to chain
def addPoint(drawsDF, q_proposed, acceptanceProb):
if np.random.binomial(1, acceptanceProb):
drawsDF.loc[drawsDF.index[-1]+1] = q_proposed ## move to proposed point
else:
q = drawsDF.iloc[-1].to_numpy()
drawsDF.loc[drawsDF.index[-1]+1] = q_current ## stay at same point
addPoint(qDF, q_proposed, acceptanceProb)
print(qDF) winningsMultiplier maxWinnings
0 0.990000 99000.00000
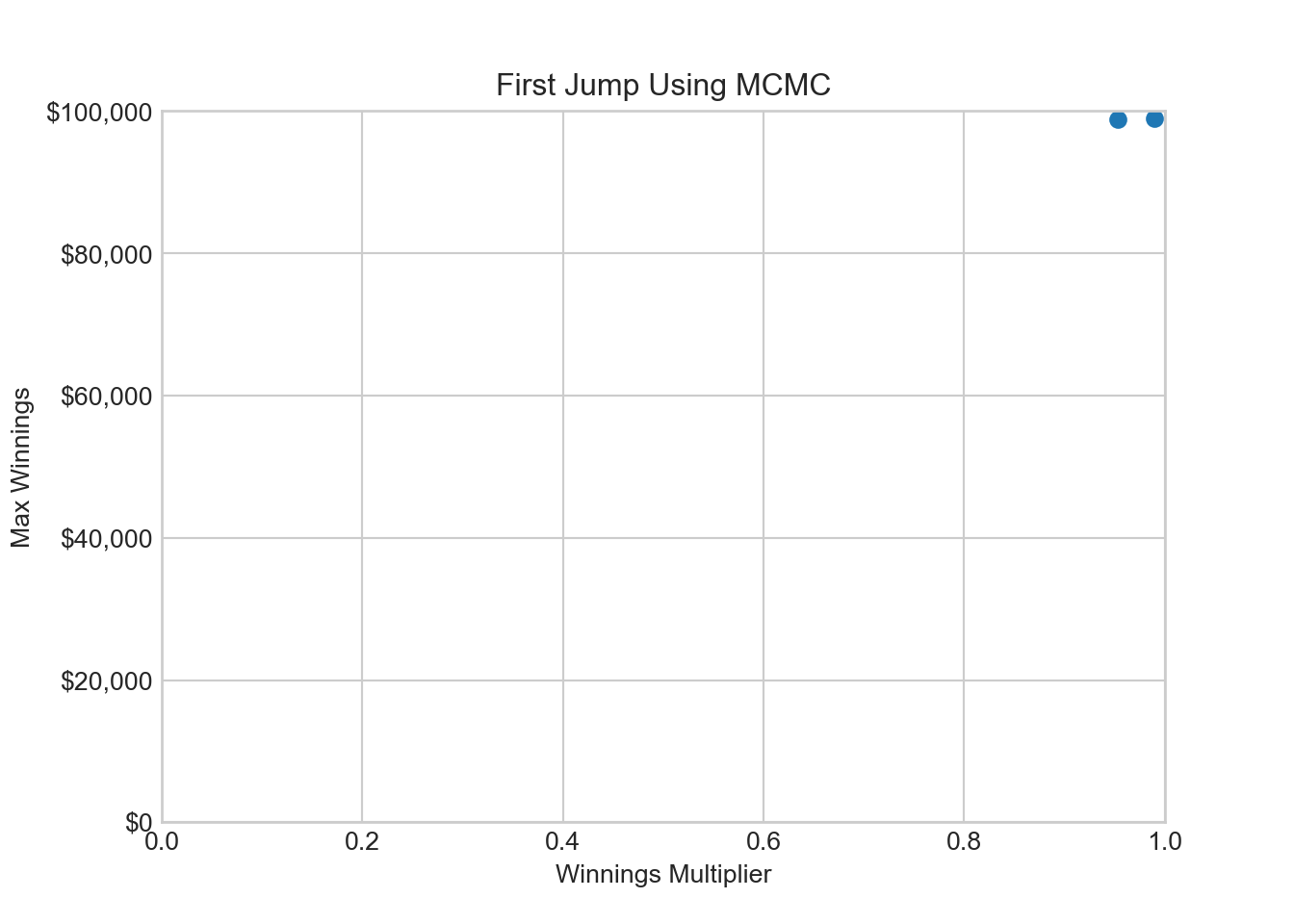
1 0.953221 98802.1757315.3.7 Unofficial Step (in between 4 & 5): Visualize the Chain
# create a scatter plot
fig, ax = plt.subplots()
ax.scatter(qDF.winningsMultiplier, qDF.maxWinnings)
# set the axis labels and title
ax.yaxis.set_major_formatter(FuncFormatter(dollar_format))
ax.set_xlabel('Winnings Multiplier')
ax.set_ylabel('Max Winnings')
ax.set_title('First Jump Using MCMC')
ax.set_xlim(0, 1)(0.0, 1.0)ax.set_ylim(0, 100000)(0.0, 100000.0)plt.show()Now, those two points in the upper-right hand corner of Figure 15.10 hardly look representative of joint distribution
15.3.8 Repeat Steps 2 - 4 Until Convergence
Now, we add a function to sample more points in our chain and then gather 200 more points.
for i in range(200):
jumpDist = rng.multivariate_normal(np.zeros(2), np.diag([0.1**2, 200**2]))
q_current = qDF.iloc[i].to_numpy()
q_proposed = q_current + jumpDist
acceptanceProb = min(1, pi_Q(q_proposed) / pi_Q(q_current))
addPoint(qDF, q_proposed, acceptanceProb)# create a scatter plot
fig, ax = plt.subplots()
ax.scatter(qDF.winningsMultiplier, qDF.maxWinnings, s= 12, alpha = 0.4)
# set the axis labels and title
ax.yaxis.set_major_formatter(FuncFormatter(dollar_format))
ax.set_xlabel('Winnings Multiplier')
ax.set_ylabel('Max Winnings')
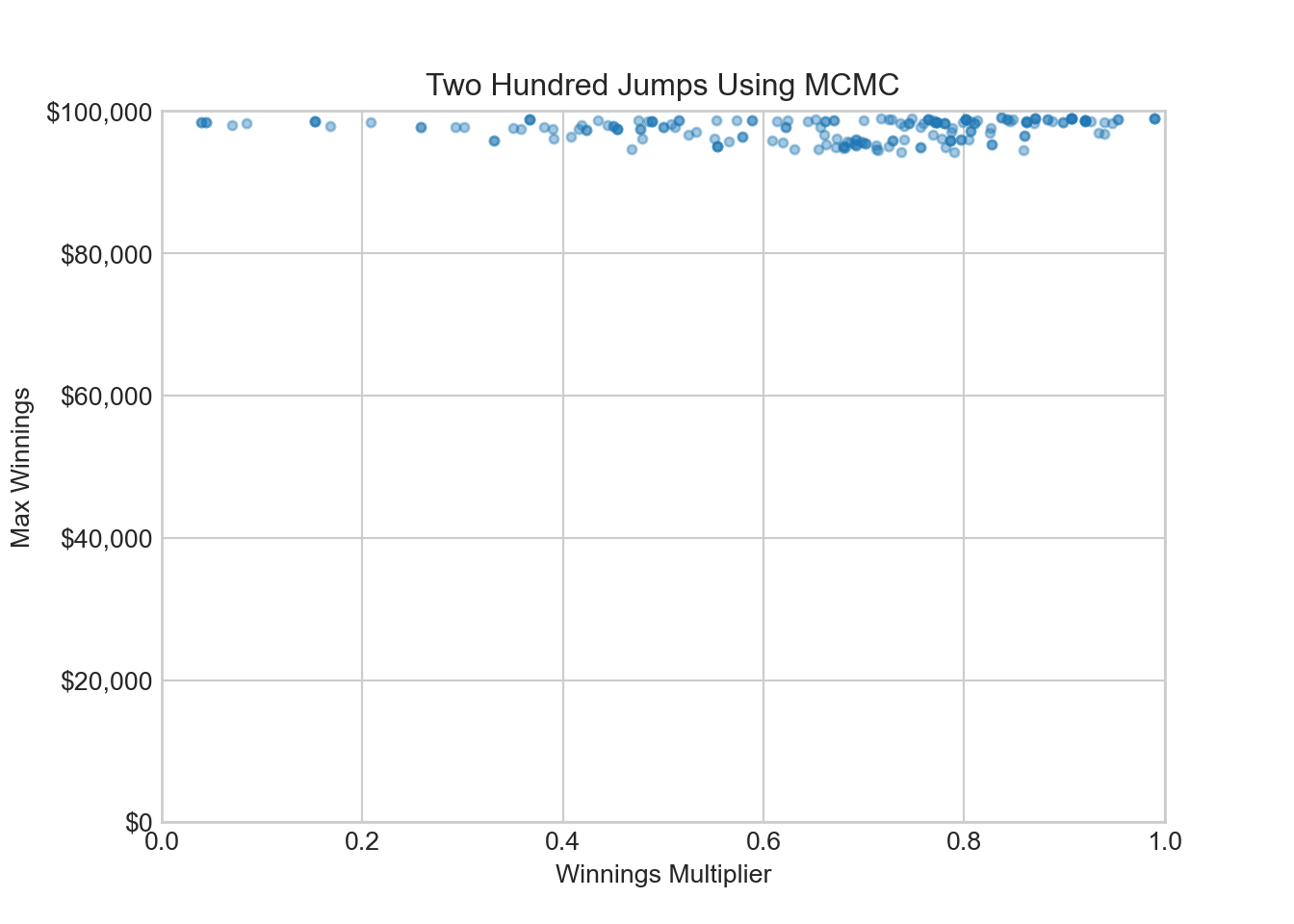
ax.set_title('Two Hundred Jumps Using MCMC')
ax.set_xlim(0, 1)(0.0, 1.0)ax.set_ylim(0, 100000)(0.0, 100000.0)plt.show()This is a fascinating plot. I will save the data associated with it as qFirstTryDF for later use.
qFirstTryDF = qDFAfter 200 jumps around parameter space, we still have not explored the sweetspot of winnings multipliers around 50% and maximum winnings around $25,000. The sample is stuck in the very flat part of the marginal density for maximum winnings
So far this correlated sample with its fancy Markov chain name seems like junk. What strategies can be employed so that our sample looks more like a representative sample from these two distributions?
15.4 Strategies to Get Representative Samples
15.4.1 More samples with bigger jumps
Part of the reason we are not finding the sweetspot of probability - often called the typical set - is that our exploration of
qDF = pd.DataFrame({
'winningsMultiplier': [0.99],
'maxWinnings': [99000.0]
})
rng = default_rng(123)
for i in range(4000): ## change to 4000 draws instead of 200
## change std dev below to 5000
jumpDist = rng.multivariate_normal(np.zeros(2), np.diag([0.1**2, 5000**2]))
q_current = qDF.iloc[i].to_numpy()
q_proposed = q_current + jumpDist
acceptanceProb = min(1, pi_Q(q_proposed) / pi_Q(q_current))
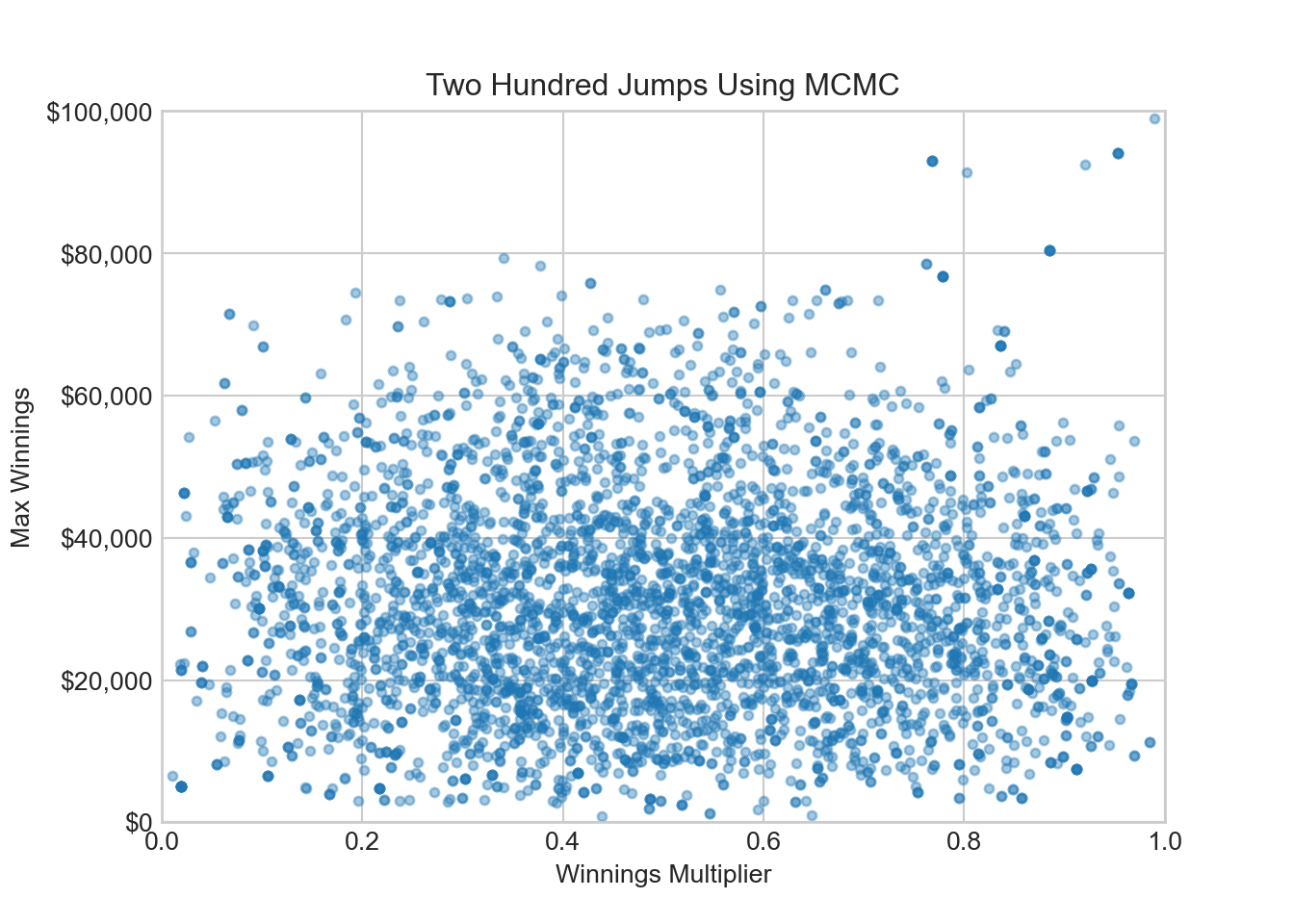
addPoint(qDF, q_proposed, acceptanceProb)After 4,000 bigger jumps around, our sampler has unstuck itself from the low-probability density region of sample space.
15.4.2 Get rid of the burn-in period
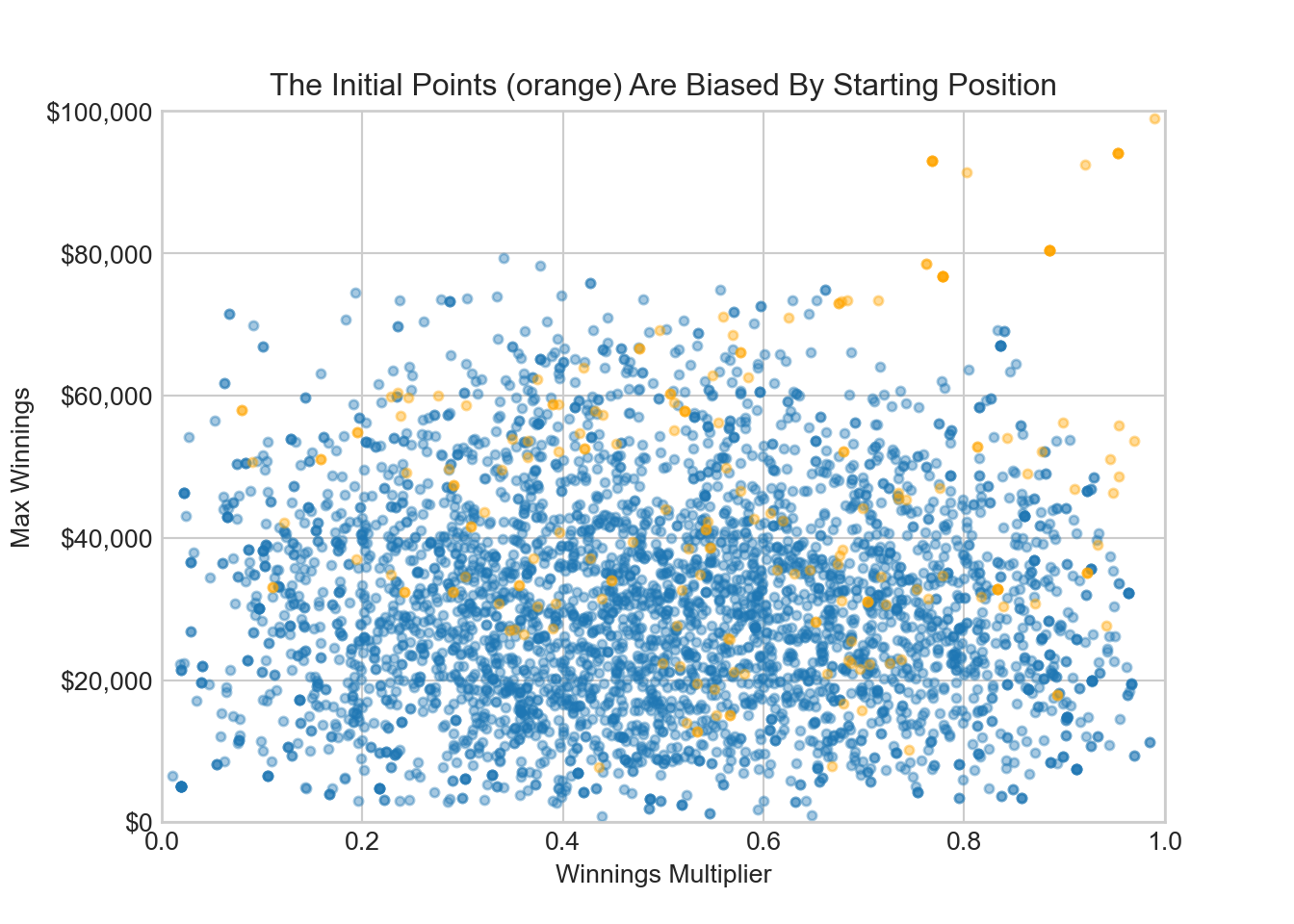
One intuition you might have is that the starting point of your sampling should not bias your results. Think of the Markov chain as following a blood hound that visits points in proportion to their joint density magnitude. If you start the blood hound’s search in a region of very low density, then a disproportionate amount of time is spent investigating points there. To eliminate this bias, a burn-in period (say first 200 samples) is discarded and only the remaining samples are used. You can see below that discarding the first 200 points removes many points where maximum winnings samples above $60,000; we started above $60,000 and that is why there is an unrepresentatively high amount of samples from that area in the burn-in samples.
15.4.3 Thinning
One last strategy to employ is called thinning. The points are correlated by design, but if we only peek at the chain every
15.5 Simple Diagnostic Checks
15.5.1 Trace Plot
Recall that our goal was to estimate expected winnings. We have previously narrowed down expected winnings to be just under $15,000. Checking this on our thinned data frame sample, we find.
(qDF.winningsMultiplier[200::3] * qDF.maxWinnings[200::3]).mean()15109.227401450673Not quite the accuracy we were hoping for, but then again we only retained 600 samples. Let’s use alot more samples, say 40,000, remove the first 1,000 as burn-in, and then retain every third sample; a total of 13,000 samples.
qDF = pd.DataFrame({
'winningsMultiplier': [0.5],
'maxWinnings': [25000.0]
})
rng = default_rng(123)
for i in range(40000): ## change to 40000 draws
## change std dev below to 1000
jumpDist = rng.multivariate_normal(np.zeros(2), np.diag([0.1**2, 5000**2]))
q_current = qDF.iloc[i].to_numpy()
q_proposed = q_current + jumpDist
acceptanceProb = min(1, pi_Q(q_proposed) / pi_Q(q_current))
addPoint(qDF, q_proposed, acceptanceProb)
## expected winnings
(qDF.winningsMultiplier[1001::3] * qDF.maxWinnings[1001::3]).mean()15288.8086311268which is a much more accurate approximation. It is good to know this works!
One of the best ways to check that your exploring parameter space properly is to use trace plots.
For our two variables, here is a trace plot of the first 2,000 draws without removing the burn-in period.
# create a scatter plot
qDF = pd.DataFrame({
'winningsMultiplier': [0.99],
'maxWinnings': [99000.0]
})
rng = default_rng(123)
for i in range(2000): ## change to 40000 draws
## change std dev below to 1000
jumpDist = rng.multivariate_normal(np.zeros(2), np.diag([0.1**2, 5000**2]))
q_current = qDF.iloc[i].to_numpy()
q_proposed = q_current + jumpDist
acceptanceProb = min(1, pi_Q(q_proposed) / pi_Q(q_current))
addPoint(qDF, q_proposed, acceptanceProb)
fig, ax = plt.subplots()
ax.scatter(qDF.winningsMultiplier[200:], qDF.maxWinnings[200:], s= 12, alpha = 0.4)
ax.scatter(qDF.winningsMultiplier[:200], qDF.maxWinnings[:200], s= 12, alpha = 0.4, c = "orange")
# set the axis labels and title
ax.yaxis.set_major_formatter(FuncFormatter(dollar_format))
ax.set_xlabel('Winnings Multiplier')
ax.set_ylabel('Max Winnings')
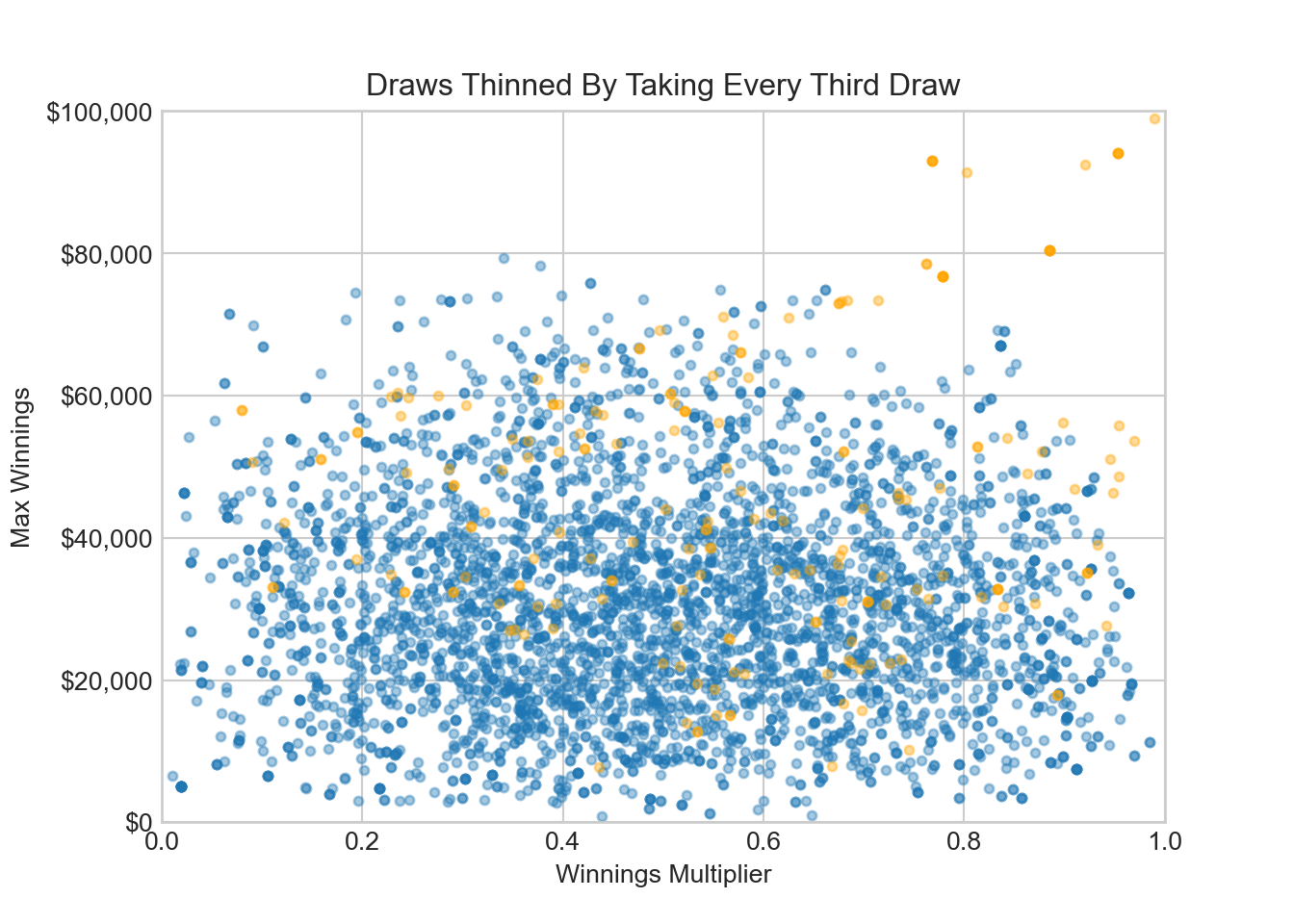
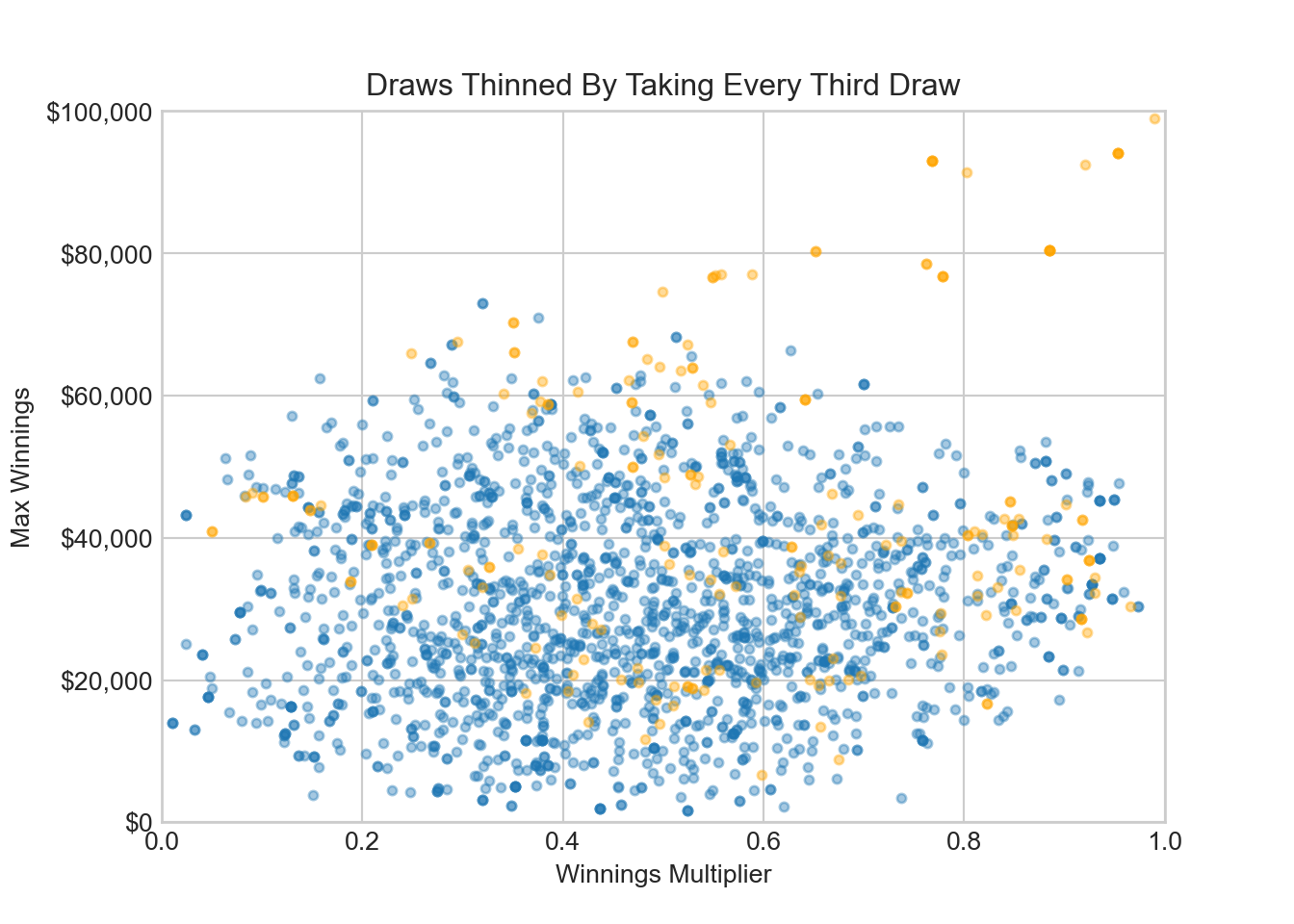
ax.set_title('Draws Thinned By Taking Every Third Draw')
ax.set_xlim(0, 1)ax.set_ylim(0, 100000)plt.show()The plot shows a lack of exploration of parameter space. The bloodhound is following probability density towards the probability sweetspot, but we can tell from the plot that parameter space exploration is insufficient at this point - there is no fuzzy caterpillar, more of a worm. In our diagnostics, we will use arviz functionality to give a trace plot. You can try this code to reproduce a fuzzy caterpillar trace plot:
import numpyro
import numpyro.distributions as dist
from jax import random
import arviz as az
def model(height, weight):
# Define priors on model parameters
beta = numpyro.sample('beta', dist.Normal(0, 10))
sigma = numpyro.sample('sigma', dist.Exponential(1))
# Define likelihood of observed data
mu = beta * height
numpyro.sample('obs', dist.Normal(mu, sigma), obs=weight)
# Generate some fake data
true_beta = 2.5
true_sigma = 1.0
num_samples = 100
height = dist.Normal(70, 2).sample(random.PRNGKey(0), num_samples)
weight = dist.Normal(true_beta * height, true_sigma).sample(random.PRNGKey(1))
# Fit the model using MCMC
from numpyro.infer import MCMC, NUTS
nuts_kernel = NUTS(model)
mcmc = MCMC(nuts_kernel, num_warmup=500, num_samples=1000)
mcmc.run(random.PRNGKey(2), height=height, weight=weight)
# Use ArviZ to plot the trace plot of the beta parameter
az.plot_trace(mcmc.get_samples()['beta'])15.6 Summary
In Monte Carlo integration, we use random sampling to estimate the value of an integral of a function over a given domain. The basic idea is to generate a large number of random points within the domain and use them to calculate an approximate value of the integral.
However, in many cases, generating independent random samples may not be feasible or efficient, especially in high-dimensional or complex problems where the domain of integration is large or the function being integrated is difficult to evaluate. In such cases, Markov Chain Monte Carlo (MCMC) methods can be used to generate a sequence of correlated samples from a probability distribution that is proportional to the function being integrated; .
The sequence of correlated samples generated by MCMC can be used to estimate the value of the integral, based on the Law of Large Numbers. As the number of samples in the sequence increases, the estimate of the integral becomes more accurate. Moreover, the correlation between successive samples ensures that the samples are drawn efficiently from the regions of the distribution that contribute most to the integral, leading to a more efficient estimation of the integral compared to simple random sampling.
Therefore, by using MCMC to generate a sequence of correlated samples, we can obtain a more efficient and accurate estimate of the integral compared to using independent random sampling.
15.7 Getting Help
TBD
15.8 Questions to Learn From
Exercise 15.1 Consider how you might go about adding the Kumaraswamy Distribution as a new sampler to be used with the numpyro package. This is poorly documented feature, but addressed on the numpyro discussion forum.
Your goal for this exercise is to create a new distribution, the Kumaraswamy distribution, for use with the numpyro. Write an .ipynb file demonstrating how you can create and use (small toy problem) a Kumaraswamy distribution.
Exercise 15.2 With increased problem complexity, increasing sampling efficiency or decreasing estimator variance becomes important to solving problems - like generating Bayesian posteriors - in a reasonable amount of time. To quickly get a feel for estimator variance, I will often do 20 simulation runs of
Calculate the expected winnings using the assumption of the spinner game above. Do this 20 times for each of the three combinations of density functions with .ipynb file.
Exercise 15.3 In proofs A.20 and A.21 of https://cs.dartmouth.edu/wjarosz/publications/dissertation/appendixA.pdf, explain what mathematical concept is being used to go from each line of the proof to the subsequent line of the proof. Upload a copy of the two proofs annotated with the mathematical concept being applied in each line as a PDF file. Example mathematical concepts include “law of total probability” or “sum of the variances equals variance of the sums for independent random variables”.
Exercise 15.4 There is code above that looks like this:
qDF = pd.DataFrame({
'winningsMultiplier': [0.5],
'maxWinnings': [25000.0]
})
rng = default_rng(123)
for i in range(40000): ## change to 40000 draws
## change std dev below to 1000
jumpDist = rng.multivariate_normal(np.zeros(2), np.diag([0.1**2, 5000**2]))
q_current = qDF.iloc[i].to_numpy()
q_proposed = q_current + jumpDist
acceptanceProb = min(1, pi_Q(q_proposed) / pi_Q(q_current))
addPoint(qDF, q_proposed, acceptanceProb)
## expected winnings
(qDF.winningsMultiplier[1001::3] * qDF.maxWinnings[1001::3]).mean()It is very slow code due to the for loop. Rewrite the code using numpy arrays and vectorized functions to make it faster. Comment on the speed improvement.
Exercise 15.5 Using the notation of this chapter (e.g. .ipynb file.
Exercise 15.6 In this exercise, we will explore how the choice of proposal distribution affects sampling efficiency. Specifically, create three different proposal distributions by adjusting the covariance matrix of the proposal distribution. The three distributions should represent these three scenarios:
- The Teleporting Couch Potato Scenario: The proposal distribution leads to roughly 5% of all proposed jumps being accepted.
- The Smart Explorer Scenario: The proposal distribution leads to roughly 50% of all proposed jumps being accepted.
- The Turtle Scenario: proposal distribution leads to roughly 99% of all proposed jumps being accepted.
Using a 10,000 sample chain and discarding the first 2,000 samples as burn-in, create a trace plot for winnings (i.e. winningsMultiplier * maxWinnings) for each of the three scenarios above.